Code
library(tidyverse)
library(ggplot2)
knitr::opts_chunk$set(echo = TRUE)Neha Jhurani
April 12, 2023
New names:
• `` -> `...2`
• `` -> `...3` Total cost of foodborne illness estimates for 15 leading foodborne pathogens
Length:27
Class :character
Mode :character
...2 ...3
Length:27 Length:27
Class :character Class :character
Mode :character Mode :character [1] "Total cost of foodborne illness estimates for 15 leading foodborne pathogens"
[2] "...2"
[3] "...3" [1] "pathogens" "case" "cost" #Reading the dataset, shows that there are a lot of rows which don't contain any values or are NA, so, I am removing those rows. The last row is the total of the 15 pathogens, so we can remove that as well.
cost_of_illness_data<- na.omit(cost_of_illness_data)
cost_of_illness_data<- cost_of_illness_data[-16,]
# Making the values numeric so that it can be plotted
cost_of_illness_data$case<-as.numeric(cost_of_illness_data$case)
cost_of_illness_data$cost<-as.numeric(cost_of_illness_data$cost)
cost_of_illness_data# A tibble: 15 × 3
pathogens case cost
<chr> <dbl> <dbl>
1 Campylobacter spp. (all species) 8.45e5 2.18e9
2 Clostridium perfringens 9.66e5 3.84e8
3 Cryptosporidium spp. (all species) 5.76e4 5.84e7
4 Cyclospora cayetanensis 1.14e4 2.57e6
5 Listeria monocytogenes 1.59e3 3.19e9
6 Norovirus 5.46e6 2.57e9
7 Salmonella (non-typhoidal species) 1.03e6 4.14e9
8 Shigella (all species) 1.31e5 1.59e8
9 Shiga toxin-producing Escherichia coli O157 (STEC O157) 6.32e4 3.11e8
10 non-O157 Shiga toxin-producing Escherichia coli (STEC non-O157) 1.13e5 3.17e7
11 Toxoplasma gondii 8.67e4 3.74e9
12 Vibrio parahaemolyticus 3.47e4 4.57e7
13 Vibrio vulnificus 9.6 e1 3.59e8
14 Vibrio non-cholera species other than V. parahaemolyticus and … 1.76e4 8.17e7
15 Yersinia enterocolitica 9.77e4 3.13e8`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.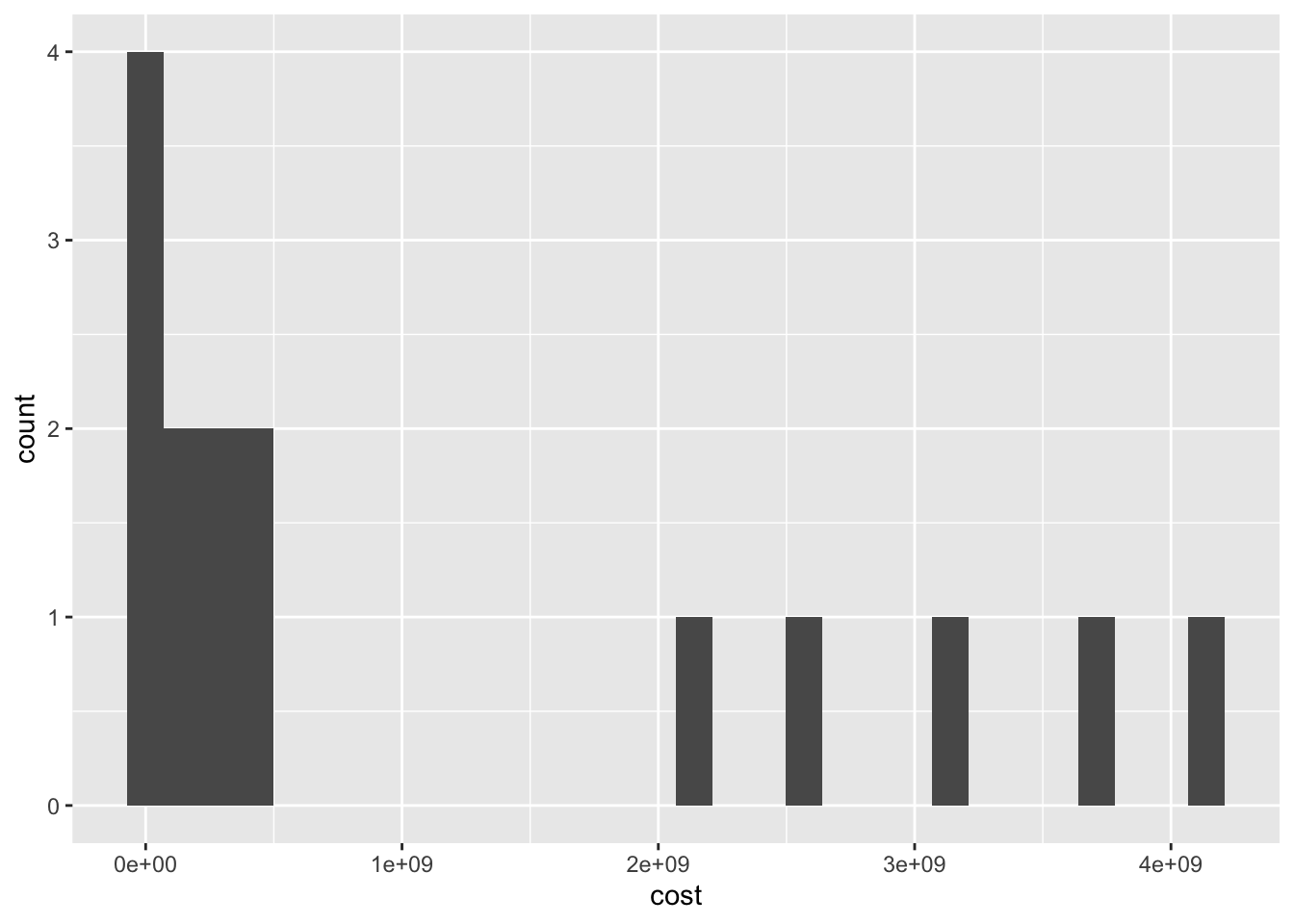
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.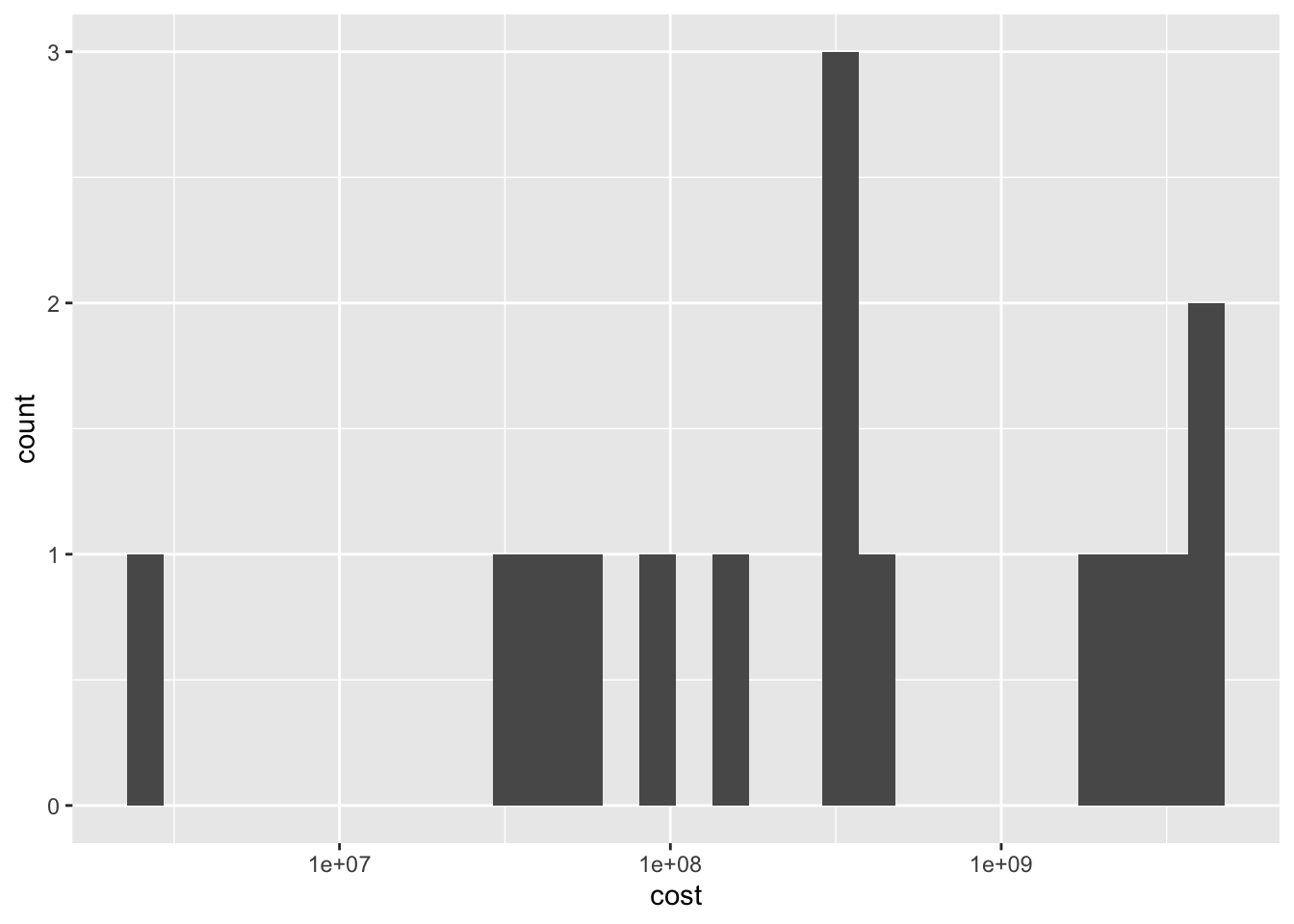
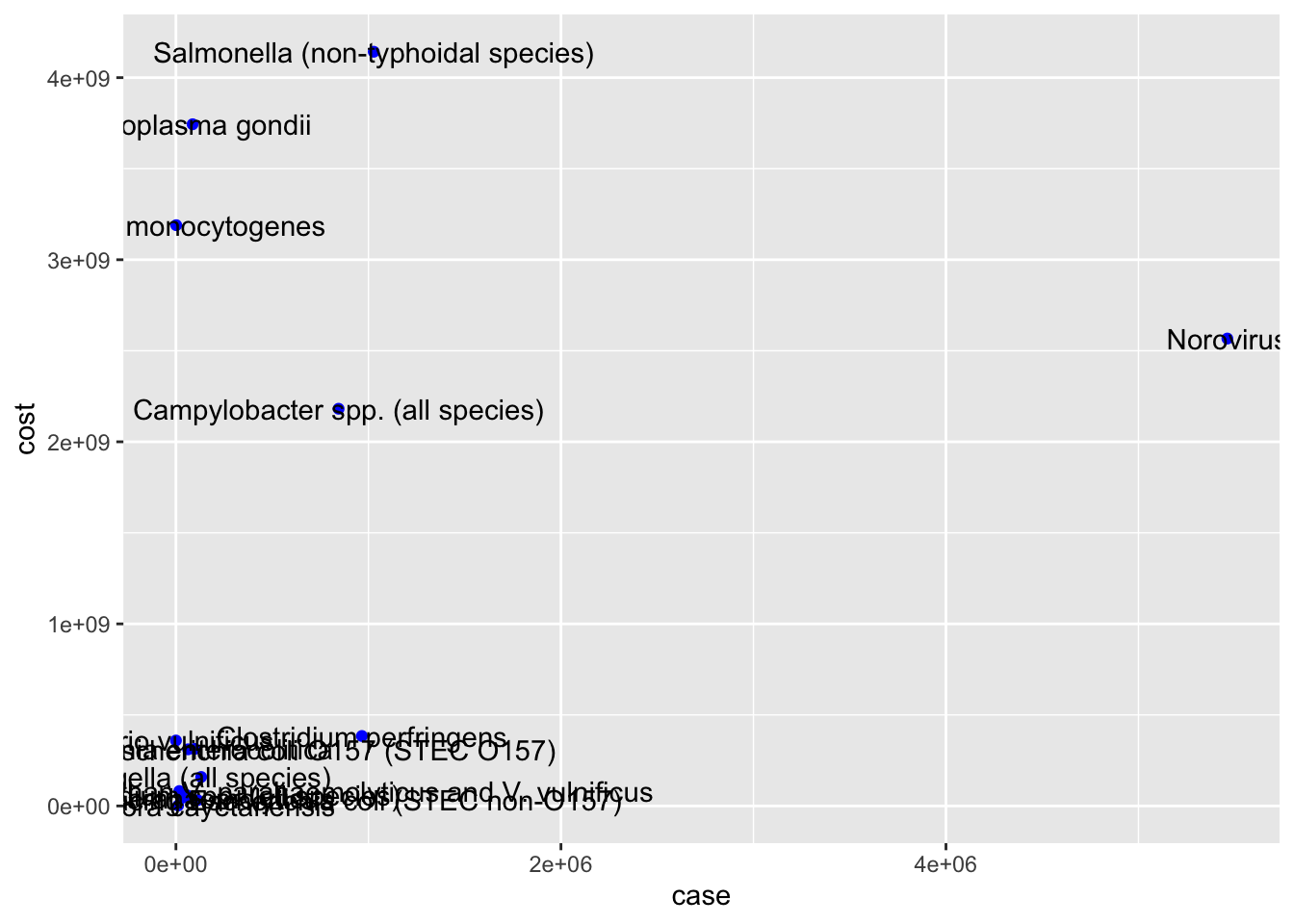
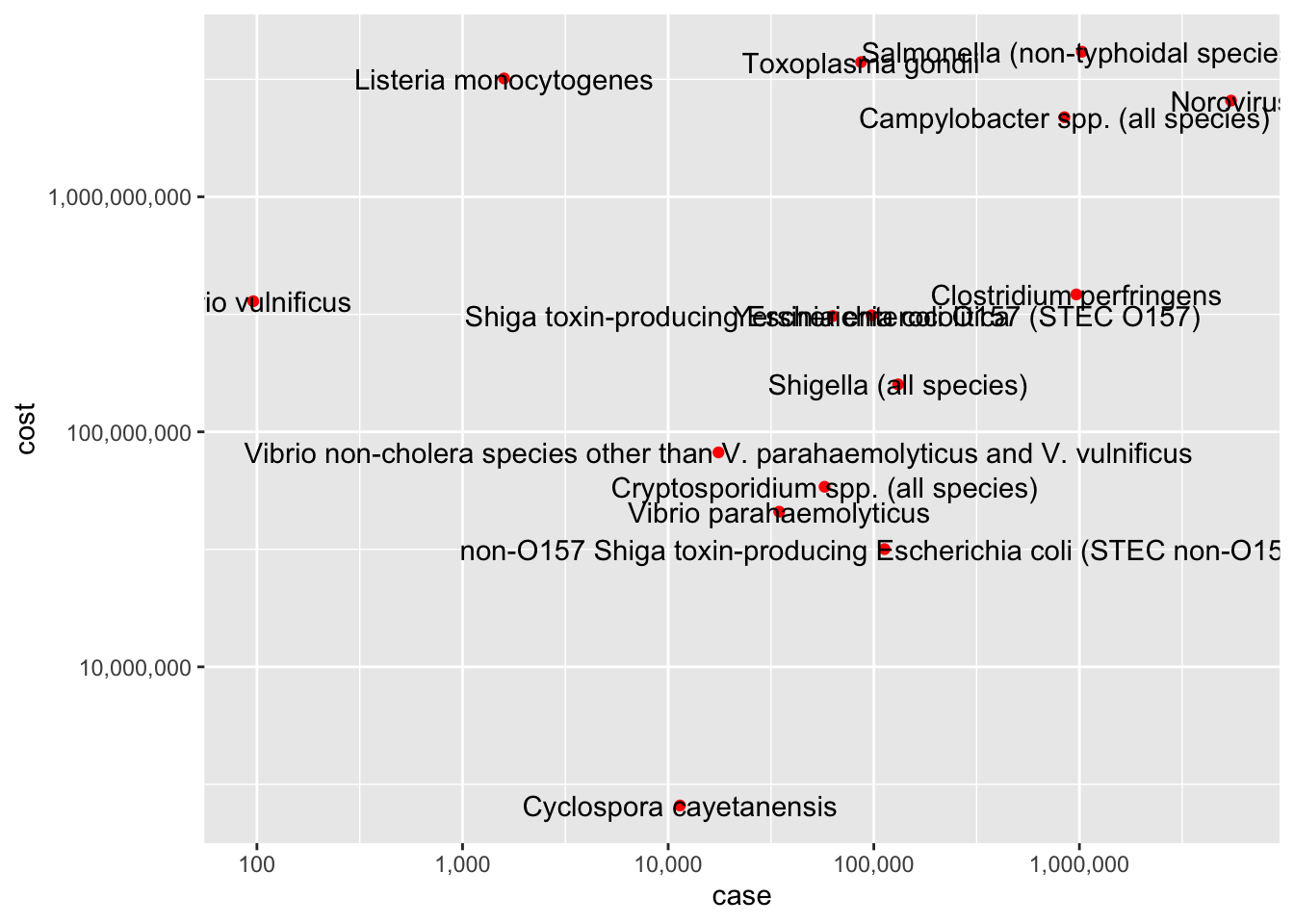
Warning: Continuous x aesthetic
ℹ did you forget `aes(group = ...)`?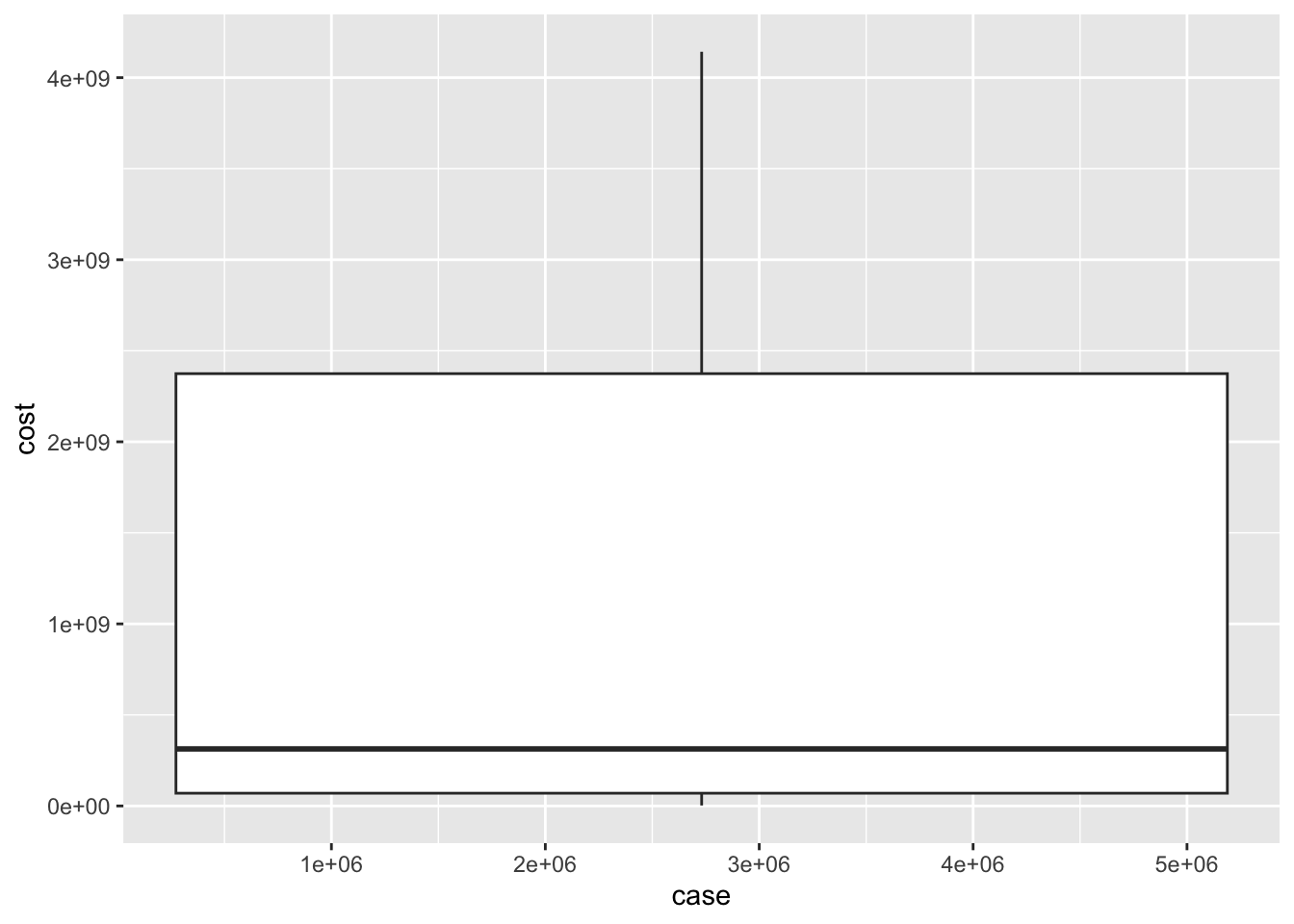
---
title: "Visualizing a dataset"
author: "Neha Jhurani"
desription: "Using ggplot2 to visualize: Total_cost_for_top_15_pathogens_2018.xlsx"
date: "04/12/2023"
format:
html:
toc: true
code-fold: true
code-copy: true
code-tools: true
categories:
- challenge5
- Neha Jhurani
- Total_cost_for_top_15_pathogens_2018.xlsx
---
```{r}
#| label: setup
#| warning: false
library(tidyverse)
library(ggplot2)
knitr::opts_chunk$set(echo = TRUE)
```
## Total_cost_for_top_15_pathogens_2018
```{r}
library(readxl)
#reading Total_cost_for_top_15_pathogens_2018 csv data
cost_of_illness_data <- read_excel("_data/Total_cost_for_top_15_pathogens_2018.xlsx")
summary(cost_of_illness_data)
#The data shows the cost associated with foodborne diseases in 2018 for top 15 foodborne pathogens causing the disease.
#extracting all the column names
colnames(cost_of_illness_data)
#Changing the column names to make it more informative and effective
colnames(cost_of_illness_data)[1] ="pathogens"
colnames(cost_of_illness_data)[2] ="case"
colnames(cost_of_illness_data)[3] ="cost"
colnames(cost_of_illness_data)
#Reading the dataset, shows that there are a lot of rows which don't contain any values or are NA, so, I am removing those rows. The last row is the total of the 15 pathogens, so we can remove that as well.
cost_of_illness_data<- na.omit(cost_of_illness_data)
cost_of_illness_data<- cost_of_illness_data[-16,]
# Making the values numeric so that it can be plotted
cost_of_illness_data$case<-as.numeric(cost_of_illness_data$case)
cost_of_illness_data$cost<-as.numeric(cost_of_illness_data$cost)
cost_of_illness_data
#Univariate Visualizations - Using the cost columns
ggplot(cost_of_illness_data, aes(x=cost)) +
geom_histogram()
ggplot(cost_of_illness_data, aes(x=cost)) +
geom_histogram()+
scale_x_continuous(trans = "log10")
# Bivariate Visualization
#x axis - number of cases, y axis - estimated costs, the points denotes the pathogens associated with it
ggplot(cost_of_illness_data, aes(case,cost,label=pathogens)) +
geom_point(color="blue")+
geom_text()
ggplot(cost_of_illness_data, aes(x=case, y=cost, label=pathogens)) +
geom_point(color = "red")+
scale_x_continuous(trans = "log10", labels = scales::comma)+
scale_y_continuous(trans = "log10", labels = scales::comma)+
geom_text()
ggplot(cost_of_illness_data, aes(case, cost)) + geom_boxplot()
```