Code
library(tidyverse)
library(readxl)
library(ggplot2)
knitr::opts_chunk$set(echo = TRUE, warning=FALSE, message=FALSE)
setwd("D:/MyDocs/Class Slides/DACSS601/601_Spring_2023/posts/_data")I was trying to analyze the ‘wild_bird_data.xlsx’ dataset in order to determine the subject of the dataset. This dataset summarizes the population size of wild bird species based on their body weight. We first start by importing the necessary libraries and setting the working directory to point to the location where the spreadsheet is located.
Notice that the first line in the spreadsheet is referring to where data is obtained from. This line is removed and the data is summarized for both the columns.Notice that the mean is a lot lesser than the maximum value for both the columns indicating that there are some very large value outliers in the dataset. The data is also left skewed as there are a lot of values clustered around the left. The mean is higher than the median because the few outliers pull the mean up.
# A tibble: 146 × 2
`Wet body weight [g]` `Population size`
<dbl> <dbl>
1 5.46 532194.
2 7.76 3165107.
3 8.64 2592997.
4 10.7 3524193.
5 7.42 389806.
6 9.12 604766.
7 8.04 192361.
8 8.70 250452.
9 8.89 16997.
10 9.52 595.
# … with 136 more rows Min. 1st Qu. Median Mean 3rd Qu. Max.
5 1821 24353 382874 198515 5093378 Min. 1st Qu. Median Mean 3rd Qu. Max.
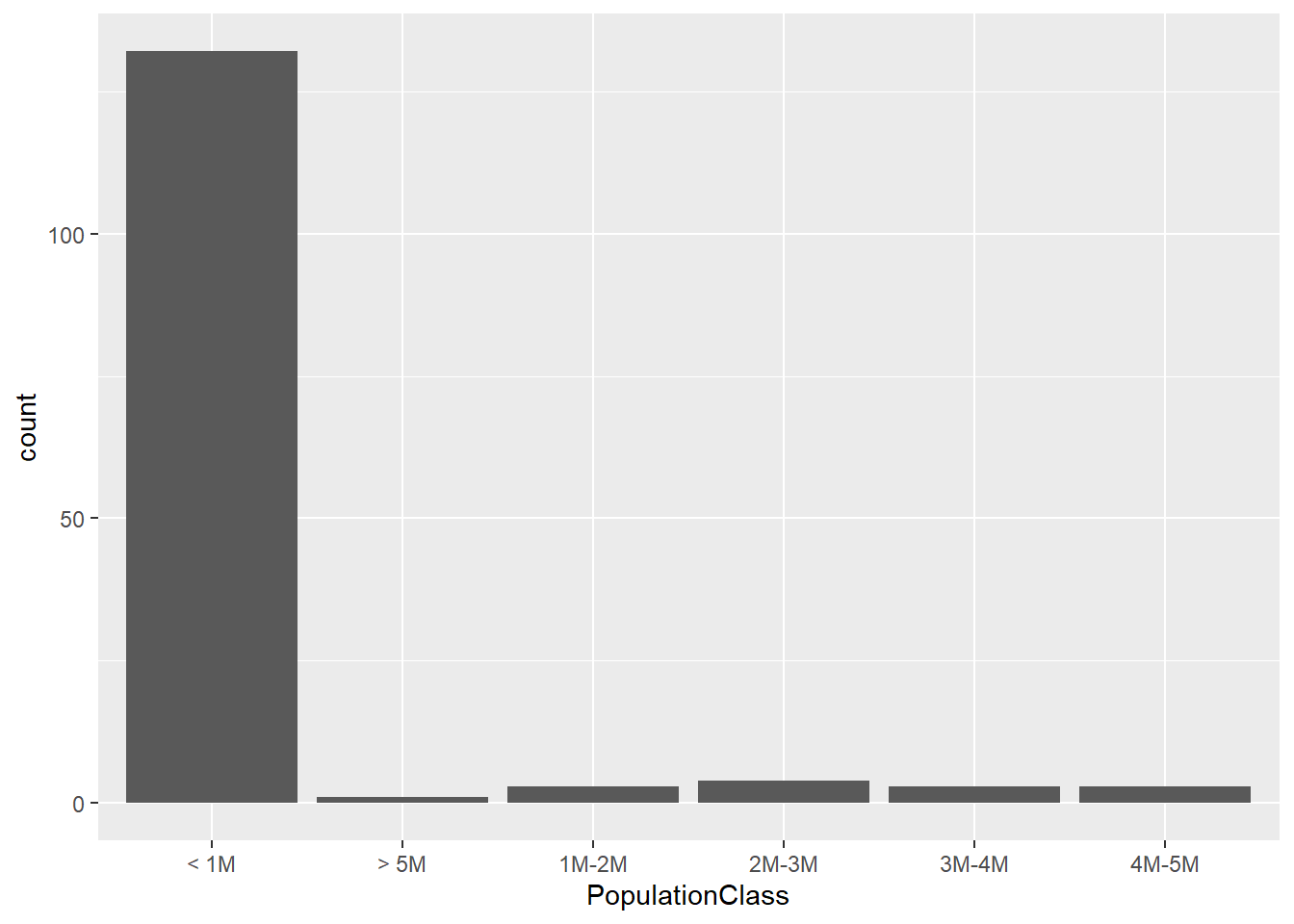
5.459 18.620 69.232 363.694 309.826 9639.845 First sorted the data according to the individual body weight and then assign each to a WeightClass in order to understand which category each bird falls into. Also assigned each bird to a PopulationClass based on Population Size. Plotted graphs which further confirm the skewness.
dataframe <- arrange(dataframe, `Wet body weight [g]`)
dataframe <- mutate(dataframe, WtClass = case_when(
`Wet body weight [g]` <= 10 ~ "Flyweight",
`Wet body weight [g]` > 10 & `Wet body weight [g]` <= 30 ~ "Bantamweight",
`Wet body weight [g]` > 30 & `Wet body weight [g]` <= 60 ~ "Featherweight",
`Wet body weight [g]` > 60 & `Wet body weight [g]` <= 100 ~ "Lightweight",
`Wet body weight [g]` > 100 & `Wet body weight [g]` <= 300 ~ "Welterweight",
`Wet body weight [g]` > 300 & `Wet body weight [g]` <= 600 ~ "Middleweight",
`Wet body weight [g]` > 600 & `Wet body weight [g]` <= 1000 ~ "Light HeavyWeight",
`Wet body weight [g]` > 1000 ~ "Heavyweight"
))
dataframe <- mutate(dataframe, PopulationClass = case_when(
`Population size` <= 1000000 ~ "< 1M",
`Population size` > 1000000 & `Population size` <= 2000000 ~ "1M-2M",
`Population size` > 2000000 & `Population size` <= 3000000 ~ "2M-3M",
`Population size` > 3000000 & `Population size` <= 4000000 ~ "3M-4M",
`Population size` > 4000000 & `Population size` <= 5000000 ~ "4M-5M",
`Population size` > 5000000 ~ "> 5M",
))
print(dataframe)# A tibble: 146 × 4
`Wet body weight [g]` `Population size` WtClass PopulationClass
<dbl> <dbl> <chr> <chr>
1 5.46 532194. Flyweight < 1M
2 7.42 389806. Flyweight < 1M
3 7.76 3165107. Flyweight 3M-4M
4 8.04 192361. Flyweight < 1M
5 8.64 2592997. Flyweight 2M-3M
6 8.70 250452. Flyweight < 1M
7 8.89 16997. Flyweight < 1M
8 9.12 604766. Flyweight < 1M
9 9.52 595. Flyweight < 1M
10 10.1 74386. Bantamweight < 1M
# … with 136 more rows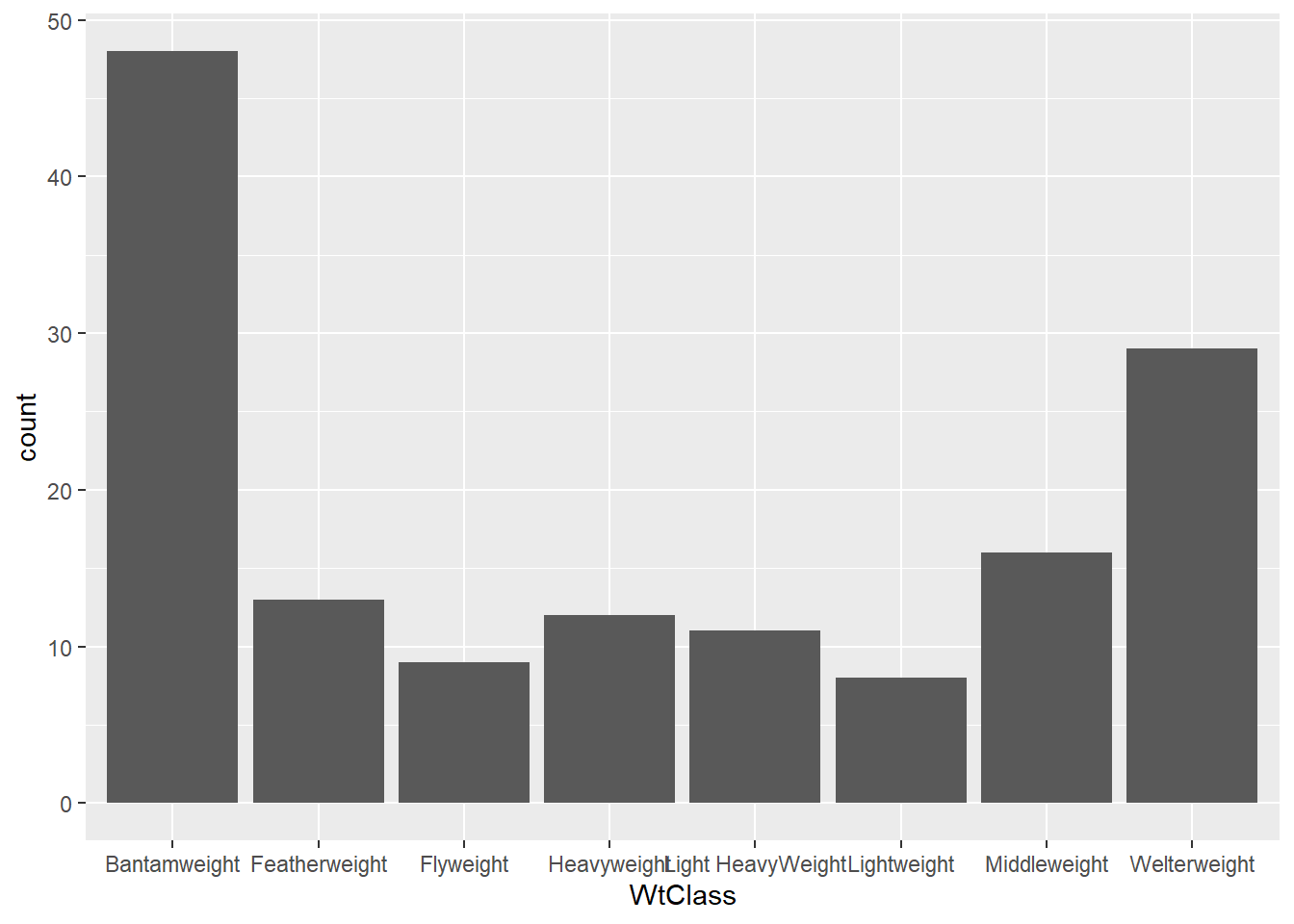
The wild_bird_data.xlsx contains information about wild bird species based on individual body weight and Population size
---
tsitle: "Categorizing Wild Bird population size based on their body weight"
author: "Sahan Prasad Podduturi Reddy"
desription: "Reading in data and creating a post"
date: "02/19/2023"
format:
html:
toc: true
code-fold: true
code-copy: true
code-tools: true
categories:
- challenge_1
- wild_bird_data.xlsx
- Sahan Prasad Podduturi Reddy
---
## Introduction
I was trying to analyze the 'wild_bird_data.xlsx' dataset in order to determine the subject of the dataset. This dataset summarizes the population size of wild bird species based on their body weight. We first start by importing the necessary libraries and setting the working directory to point to the location where the spreadsheet is located.
```{r}
#| label: Introduction
#| warning: false
#| message: false
library(tidyverse)
library(readxl)
library(ggplot2)
knitr::opts_chunk$set(echo = TRUE, warning=FALSE, message=FALSE)
setwd("D:/MyDocs/Class Slides/DACSS601/601_Spring_2023/posts/_data")
```
## Clean file and Obtain Summary Statistics
Notice that the first line in the spreadsheet is referring to where data is obtained from. This line is removed and the data is summarized for both the columns.Notice that the mean is a lot lesser than the maximum value for both the columns indicating that there are some very large value outliers in the dataset. The data is also left skewed as there are a lot of values clustered around the left. The mean is higher than the median because the few outliers pull the mean up.
```{r}
#| label: ReadFile
#| warning: false
#| message: false
setwd("D:/MyDocs/Class Slides/DACSS601/601_Spring_2023/posts/_data")
dataframe <- read_excel("wild_bird_data.xlsx", skip=1)
print(dataframe)
summary(dataframe$`Population size`)
summary(dataframe$`Wet body weight [g]`)
```
## Decribe data
First sorted the data according to the individual body weight and then assign each to a WeightClass in order to understand which category each bird falls into. Also assigned each bird to a PopulationClass based on Population Size.
Plotted graphs which further confirm the skewness.
```{r}
#| label: DescribeFile
#| warning: false
#| message: false
dataframe <- arrange(dataframe, `Wet body weight [g]`)
dataframe <- mutate(dataframe, WtClass = case_when(
`Wet body weight [g]` <= 10 ~ "Flyweight",
`Wet body weight [g]` > 10 & `Wet body weight [g]` <= 30 ~ "Bantamweight",
`Wet body weight [g]` > 30 & `Wet body weight [g]` <= 60 ~ "Featherweight",
`Wet body weight [g]` > 60 & `Wet body weight [g]` <= 100 ~ "Lightweight",
`Wet body weight [g]` > 100 & `Wet body weight [g]` <= 300 ~ "Welterweight",
`Wet body weight [g]` > 300 & `Wet body weight [g]` <= 600 ~ "Middleweight",
`Wet body weight [g]` > 600 & `Wet body weight [g]` <= 1000 ~ "Light HeavyWeight",
`Wet body weight [g]` > 1000 ~ "Heavyweight"
))
dataframe <- mutate(dataframe, PopulationClass = case_when(
`Population size` <= 1000000 ~ "< 1M",
`Population size` > 1000000 & `Population size` <= 2000000 ~ "1M-2M",
`Population size` > 2000000 & `Population size` <= 3000000 ~ "2M-3M",
`Population size` > 3000000 & `Population size` <= 4000000 ~ "3M-4M",
`Population size` > 4000000 & `Population size` <= 5000000 ~ "4M-5M",
`Population size` > 5000000 ~ "> 5M",
))
print(dataframe)
ggplot(dataframe, aes(`WtClass`)) + geom_bar()
ggplot(dataframe, aes(`PopulationClass`)) + geom_bar()
```
## Conclusion
The wild_bird_data.xlsx contains information about wild bird species based on individual body weight and Population size