knitr::opts_chunk$set(echo = TRUE, warning=FALSE, message=FALSE)
library(readxl)
library(tidyverse)
library(ggplot2)
library(dplyr)
library(stringr)
library(googlesheets4)
library(plotly)
library(stargazer)Final Project
Research Question
Does political partisanship correlate with COVID-19 death rates?
Introduction
The COVID-19 pandemic became a political matter. Early in the pandemic, mask mandates were protested in some communities. Behaviors associated with COVID-19 prevention, such as masking, social distancing, and vaccine uptake, were adopted on partisan lines. My research question is have these behaviors affected COVID-19 death rates along partisan lines?
For this analysis, I used cumulative COVID-19 death toll as opposed to infection rates as infection rates are constantly changing over time. Other studies have looked at infection rates on partisan lines over waves of the pandemic, (see this study from the Pew Research Center (Jones 2022)). I measured partisanship using 2016 county-level election results (% voting for Trump). My research is looking to see if (county-level) Trump support correlates with COVID-19 death rates. The unit of analysis for this study was U.S. counties.
If there is a systematic difference, public health interventions could target communities that may be higher risk for COVID-19 deaths based on political partisanship.
I also control for age (percent of the population over 65), income (median household income in 2020), urbanization (Urban-Rural Continuum Code), and policy (2020 governor dummy variable). Later in the analysis, I add a control for vaccination rates as previous studies have found this to be the mechanism behind this relationship (Wood and Brumfiel 2021).
Hypothesis
While I came up with this research idea on my own, other organizations such as NPR (Wood and Brumfiel 2021) and the Pew Research Center (Jones 2022) have already tested this and found a significant correlation in Trump support and COVID-19 death and infection rates. For this project, I will use more recent data and include additional control variables that were not accounted for in these previous studies.
H0: B1 (the correlation coefficient for political partisanship) is zero. There is no correlation between political partisanship and COVID-19 death rates. HA: B1 is not zero. There is a correlation between partisanship and COVID-19 death rates.
Variables
Political Partisanship
For this project, I measured partisanship as percentage of the county that voted for Trump in the 2016 election against Clinton. I did not use 2020 election results in case counties “flipped” support as a result of COVID-19. Below I tidied the data, filtering out only 2016 election results, created a variable: percent voting for Trump in 2016, and selected the percent_trump and FIPS Code variables to use in the analysis.
The data used for this variable came from the MIT Election Data and Science Lab (2021).
gs4_deauth()
#2016 election df
votedf<-read_sheet("https://docs.google.com/spreadsheets/d/1fmxoA_bibvsxsvgRdVPCgMA7DkmJNZfxiWgLgCLcsOY/edit#gid=937778872")
votedf$county_fips <- as.character(votedf$county_fips)
votedf<-mutate(votedf, county_fipsNEW=str_pad(votedf$county_fips, 5, pad = "0"))
votedf<-votedf %>%
filter(year==2016, candidate=="DONALD TRUMP")
votedf$percent_trump <-votedf$candidatevotes/votedf$totalvotes
votedf<- select(votedf, county_fipsNEW, percent_trump)
head(votedf)# A tibble: 6 × 2
county_fipsNEW percent_trump
<chr> <dbl>
1 01001 0.728
2 01003 0.765
3 01005 0.521
4 01007 0.764
5 01009 0.893
6 01011 0.242COVID-19 Cumulative Death Rate
This data comes from USAFacts.org. Their collection methods are thoroughly described on their website (see USA Facts 2022 in References). Originally I planned on using CDC/NIH data, however they only had data for a little over 1,000 counties, compared to USAFacts which had data for over 3,000. USAFacts compiles their data from CDC, but also town, county, and state leading to a larger number of observations in their dataset.
Cumulative county-level death toll is taken as of March 19, 2022, when many states stopped regularly reporting COVID-19 deaths (and beginning late in January 2020) (USA Facts 2022).
coviddf_2<-read_sheet("https://docs.google.com/spreadsheets/d/1ZKa3sg_UdtyX5z0OGGVZN6nuqcC_OKWcsOjm1ZHNjQY/edit#gid=716391091")
#adding leading 0 back to fips
coviddf_2<-mutate(coviddf_2, county_fipsNEW=str_pad(coviddf_2$"countyFIPS", 5, pad = "0"))
#selecting march 19, 2022, the day before the first day of spring in 2022, when many states stopped/slowed reporting (USA Facts)
coviddf_2<-select(coviddf_2, county_fipsNEW, "County Name", State, "2022-03-19")
coviddf_2<-rename(coviddf_2, "covid_deaths" = "2022-03-19")
head(coviddf_2)# A tibble: 6 × 4
county_fipsNEW `County Name` State covid_deaths
<chr> <chr> <chr> <chr>
1 00000 Statewide Unallocated AL 0
2 01001 Autauga County AL 210
3 01003 Baldwin County AL 669
4 01005 Barbour County AL 94
5 01007 Bibb County AL 100
6 01009 Blount County AL 230 Control: Age
Because age correlates with political party and older Americans (over 65) were disproportionately more likely to die as a result of COVID-19, this is included in the analysis as a control. According to an article from the Mayo Clinic Website, 81% of COVID-19 deaths occurred in individuals 65 or over (Mayo Clinic Staff 2022). Previous studies have controlled for age as well (Brumfiel and Wood 2021). I controlled for age by creating the variable percent of the county’s population over 65 years of age.
#Please see pdf of code tidying I did to make df into a manageable size for google sheets
age<- read_sheet("https://docs.google.com/spreadsheets/d/1EysREWJ61NCSYyiYH8-2pmZC_TnuLTgqa8Lz5ZpsPZ4/edit#gid=271068707")
head(age)# A tibble: 6 × 6
STNAME CTYNAME county_fipsNEW tot_pop over65 over65_pct
<chr> <chr> <chr> <chr> <chr> <chr>
1 Alabama Autauga County 01001 55869 8924 0.1597307988329843
2 Alabama Baldwin County 01003 223234 46830 0.20977987224168362
3 Alabama Barbour County 01005 24686 4861 0.19691323017094708
4 Alabama Bibb County 01007 22394 3733 0.16669643654550326
5 Alabama Blount County 01009 57826 10814 0.18700930377338915
6 Alabama Bullock County 01011 10101 1711 0.1693891693891694 Control: Policy Dummy Variable
This variable (2020 governor party) attempts to control for local and state policy such as mask mandates, stay-at-home orders, and vaccine requirements to attend large events that contribute to COVID-19 death and infection rates. This data set was created from a table of current U.S. governors and their political party on Ballotpedia.org (2022). I adjusted a couple observations when governors had been elected more recently than March 11, when the WHO declared COVID-19 a global pandemic. This data represents the political party of state governors as of March 11, 2020.
#Governor/policy df
#reading in gov dummy variable: whoever was in office March 11, 2020 (when WHO declared covid-19 a global pandemic)
gov2020<-read_sheet("https://docs.google.com/spreadsheets/d/1-pToTikvnXl1-lT-xazjoxX0sOCwOH5-7Bl8vJqu9Tk/edit#gid=0")
gov2020$Office<-str_remove(gov2020$Office, "Governor of ")
gov2020<-rename(gov2020, "STNAME" = Office)
gov2020<-select(gov2020, STNAME, Name, Party)
head(gov2020)# A tibble: 6 × 3
STNAME Name Party
<chr> <chr> <chr>
1 Alabama Kay Ivey Republican
2 Alaska Mike Dunleavy Republican
3 American Samoa Lemanu Palepoi Mauga Democratic
4 Arizona Doug Ducey Republican
5 Arkansas Asa Hutchinson Republican
6 California Gavin Newsom DemocraticControl: Income
Median household income by county in 2020 is taken from the Economic Research Service at United States Department of Agriculture (2022). Income likely correlates with both political affiliation and COVID-19 death rates (access to medical care, preventative treatment etc), which is why it is included in the analysis.
Control: Rural-Urban Continuum Code
This is also taken from the Economic Research Service/USDA dataset. According to the USDA, the 2013 rural-urban continuum code is a “classification scheme that distinguishes metropolitan counties by the population size of their metro area, and non-metropolitan counties by degree of urbanization and adjacency to a metro area”. In my opinion, this makes more sense to include than a simple calculation of population density, because it takes into account cities/ how close people settle to cities rather than just population divided by land area. (For instance, a large county, land-wise, may have a majority of the population in a large city, however this could still result in a fairly small population density depending on land area). The Rural Urban Continuum code is on an integer scale from 1 to 9, where 1 represents the most urban/metro counties, and 9 represents the most rural counties.
Generally speaking, more rural counties tend to favor Trump while more metropolitan areas tend to be more democratic. At the same time, one would expect that more densely populated areas/ people living closer together in cities would experience higher infection (and therefore death rates) of COVID-19.
#Income and density df
#reading in income variable sheet, renaming variables, and selecting only relevant columns, then renaming fips code variable to join to other df's
income<-read_sheet("https://docs.google.com/spreadsheets/d/1ntReIIrpzjRvGabr64-91xEpSJk10r6Er1CX3pG5zBg/edit#gid=1233692484", skip=4) %>%
rename("med_income_2020" = Median_Household_Income_2020, "county_fipsNEW" = FIPS_code) %>%
select(county_fipsNEW, State, Area_name, med_income_2020, Rural_urban_continuum_code_2013)
head(income)# A tibble: 6 × 5
county_fipsNEW State Area_name med_income_2020 Rural_urban_continuu…¹
<chr> <chr> <chr> <dbl> <dbl>
1 00000 US United States 67340 NA
2 01000 AL Alabama 53958 NA
3 01001 AL Autauga County, AL 67565 2
4 01003 AL Baldwin County, AL 71135 3
5 01005 AL Barbour County, AL 38866 6
6 01007 AL Bibb County, AL 50907 1
# … with abbreviated variable name ¹Rural_urban_continuum_code_2013Joining the Dataframes
Below I join the data based on FIPS code (except for the governor policy dummy variable, which is joined by state).
covidvote_2<-votedf %>%
left_join(coviddf_2, by="county_fipsNEW")
covid_vote_3 <- covidvote_2 %>%
left_join(age, by= "county_fipsNEW")
covid_vote_4<- covid_vote_3 %>%
left_join(gov2020, by="STNAME")
covid_vote_5<- covid_vote_4 %>%
left_join(income, by= "county_fipsNEW")
head(covid_vote_5)# A tibble: 6 × 16
county…¹ perce…² Count…³ State.x covid…⁴ STNAME CTYNAME tot_pop over65 over6…⁵
<chr> <dbl> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 01001 0.728 Autaug… AL 210 Alaba… Autaug… 55869 8924 0.1597…
2 01003 0.765 Baldwi… AL 669 Alaba… Baldwi… 223234 46830 0.2097…
3 01005 0.521 Barbou… AL 94 Alaba… Barbou… 24686 4861 0.1969…
4 01007 0.764 Bibb C… AL 100 Alaba… Bibb C… 22394 3733 0.1666…
5 01009 0.893 Blount… AL 230 Alaba… Blount… 57826 10814 0.1870…
6 01011 0.242 Bulloc… AL 52 Alaba… Bulloc… 10101 1711 0.1693…
# … with 6 more variables: Name <chr>, Party <chr>, State.y <chr>,
# Area_name <chr>, med_income_2020 <dbl>,
# Rural_urban_continuum_code_2013 <dbl>, and abbreviated variable names
# ¹county_fipsNEW, ²percent_trump, ³`County Name`, ⁴covid_deaths, ⁵over65_pct#covid_vote_5 has all 5 dataframes joined togetherMore tidying:
#converting covid deaths, age, and total population to numeric variables
covid_vote_5$covid_deaths<-as.numeric(covid_vote_5$covid_deaths)
covid_vote_5$tot_pop<-as.numeric(covid_vote_5$tot_pop)
covid_vote_5$over65_pct<-as.numeric(covid_vote_5$over65_pct)
#creating a death rate variable (COVID-19 deaths per 100,000 by county from January 2020-March 2022)
covid_vote_5$"covid_death_rate" = (covid_vote_5$covid_deaths / covid_vote_5$tot_pop)*100000
#selecting only relevant columns for analysis
covid_vote_5<- select(covid_vote_5, county_fipsNEW, STNAME, CTYNAME, covid_death_rate, percent_trump, over65_pct, Party, med_income_2020, tot_pop, Rural_urban_continuum_code_2013)
head(covid_vote_5)# A tibble: 6 × 10
county_…¹ STNAME CTYNAME covid…² perce…³ over6…⁴ Party med_i…⁵ tot_pop Rural…⁶
<chr> <chr> <chr> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
1 01001 Alaba… Autaug… 376. 0.728 0.160 Repu… 67565 55869 2
2 01003 Alaba… Baldwi… 300. 0.765 0.210 Repu… 71135 223234 3
3 01005 Alaba… Barbou… 381. 0.521 0.197 Repu… 38866 24686 6
4 01007 Alaba… Bibb C… 447. 0.764 0.167 Repu… 50907 22394 1
5 01009 Alaba… Blount… 398. 0.893 0.187 Repu… 55203 57826 1
6 01011 Alaba… Bulloc… 515. 0.242 0.169 Repu… 33124 10101 6
# … with abbreviated variable names ¹county_fipsNEW, ²covid_death_rate,
# ³percent_trump, ⁴over65_pct, ⁵med_income_2020,
# ⁶Rural_urban_continuum_code_2013Removing counties with death rates of zero:
# Creating df where counties with death rates of exactly 0 (no covid deaths) are excluded. Doing this exclude counties that likely did not report COVID-19 deaths. Filtering counties where covid_death_pct is greater than zero removes 66 counties from the analysis.
covid_vote_5_no_zero<-filter(covid_vote_5, covid_death_rate >0)
head(covid_vote_5_no_zero)# A tibble: 6 × 10
county_…¹ STNAME CTYNAME covid…² perce…³ over6…⁴ Party med_i…⁵ tot_pop Rural…⁶
<chr> <chr> <chr> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
1 01001 Alaba… Autaug… 376. 0.728 0.160 Repu… 67565 55869 2
2 01003 Alaba… Baldwi… 300. 0.765 0.210 Repu… 71135 223234 3
3 01005 Alaba… Barbou… 381. 0.521 0.197 Repu… 38866 24686 6
4 01007 Alaba… Bibb C… 447. 0.764 0.167 Repu… 50907 22394 1
5 01009 Alaba… Blount… 398. 0.893 0.187 Repu… 55203 57826 1
6 01011 Alaba… Bulloc… 515. 0.242 0.169 Repu… 33124 10101 6
# … with abbreviated variable names ¹county_fipsNEW, ²covid_death_rate,
# ³percent_trump, ⁴over65_pct, ⁵med_income_2020,
# ⁶Rural_urban_continuum_code_2013#Checking data set summary
summary(covid_vote_5_no_zero) county_fipsNEW STNAME CTYNAME covid_death_rate
Length:3092 Length:3092 Length:3092 Min. : 11.38
Class :character Class :character Class :character 1st Qu.: 243.91
Mode :character Mode :character Mode :character Median : 349.97
Mean : 358.65
3rd Qu.: 458.69
Max. :1211.31
percent_trump over65_pct Party med_income_2020
Min. :0.04087 Min. :0.04859 Length:3092 Min. : 22901
1st Qu.:0.54454 1st Qu.:0.16715 Class :character 1st Qu.: 47695
Median :0.66309 Median :0.19403 Mode :character Median : 55044
Mean :0.63214 Mean :0.19766 Mean : 57333
3rd Qu.:0.74841 3rd Qu.:0.22241 3rd Qu.: 63872
Max. :0.94585 Max. :0.58174 Max. :160305
tot_pop Rural_urban_continuum_code_2013
Min. : 169 Min. :1.000
1st Qu.: 11323 1st Qu.:2.000
Median : 26308 Median :6.000
Mean : 105893 Mean :4.962
3rd Qu.: 69704 3rd Qu.:7.000
Max. :10039107 Max. :9.000 Explotatory Analysis
#Scatter plot of percent trump and percent population dying of covid.
visual1<-ggplot(covid_vote_5_no_zero, aes(x=percent_trump, y=covid_death_rate))+geom_point()+geom_smooth(size=0.5, color="firebrick")+labs(x= "Proportion Voting for Trump in 2016", y="COVID-19 Deaths per 100,000 People", title= "COVID-19 Death Rate Based on 2016 Trump Support")
ggplotly(visual1)There appears to be a slight positive trend, especially among counties where percentage Trump votes was over 50%. Interestingly, on this plot, the death rate appears to decrease slightly from 0 to 40% percent voting for Trump, and then increases. Perhaps this could be due to the fact that highly democratic counties are more likely to be more urban- therefore people live more close together and infection rate is likely higher. From this visual, it appears that the county with the highest COVID-19 death rate (1121 per 100,000) voted heavily for Trump in 2016 (over 90%).
A simple linear regression model show a slight positive trend between proportion voting for Trump and cumulative death rate.
ggplot(covid_vote_5_no_zero, aes(x=percent_trump, y=covid_death_rate))+geom_point()+geom_smooth(method= "lm", size=0.5, color="firebrick")+labs(x= "Proportion Voting for Trump in 2016", y="COVID-19 Deaths per 100,000 People", title= "COVID-19 Death Rate Based on 2016 Trump Support")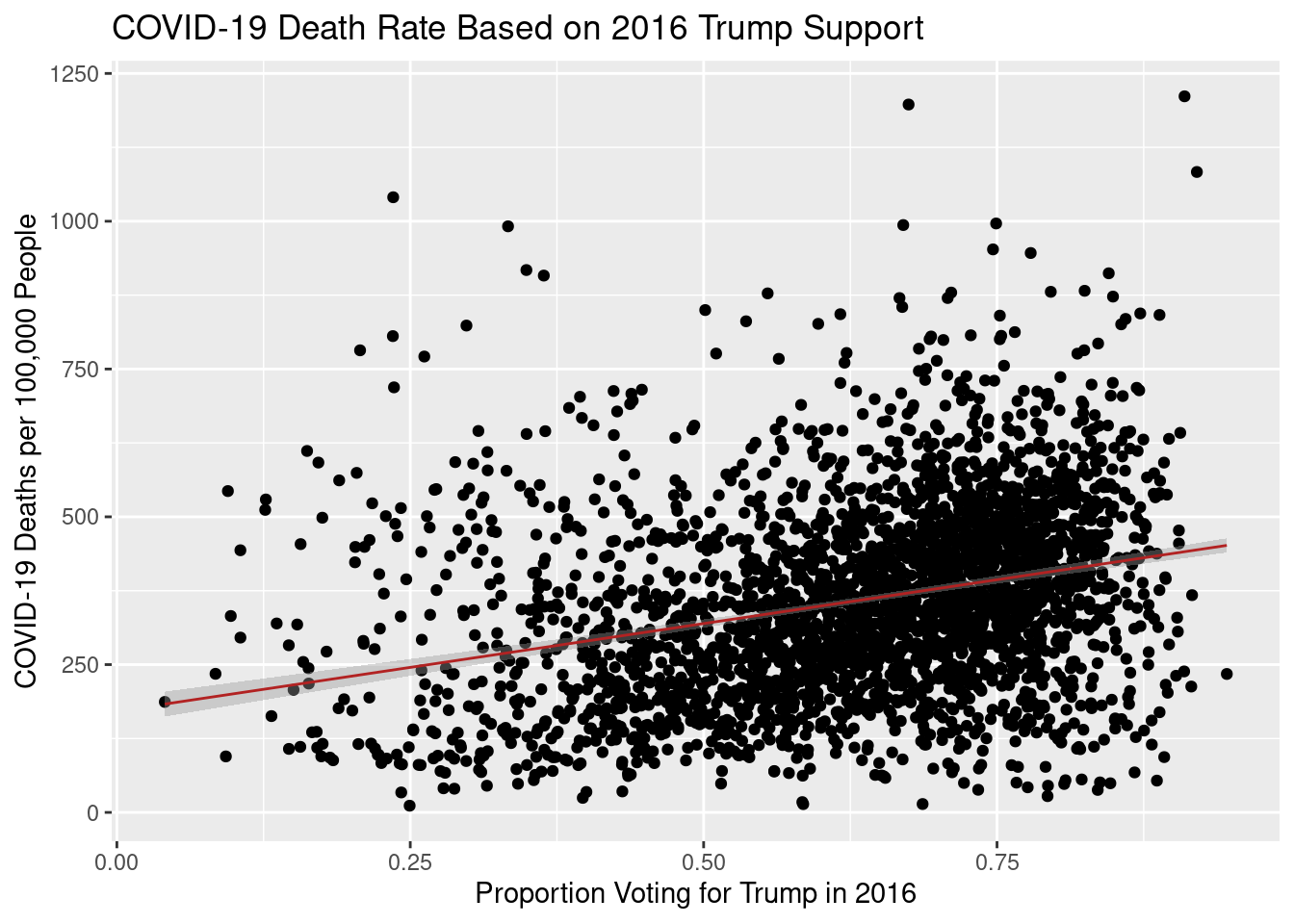
summary(lm(covid_death_rate ~ percent_trump, data= covid_vote_5_no_zero))
Call:
lm(formula = covid_death_rate ~ percent_trump, data = covid_vote_5_no_zero)
Residuals:
Min 1Q Median 3Q Max
-381.00 -104.99 -11.36 86.60 826.15
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 171.17 11.44 14.96 <2e-16 ***
percent_trump 296.58 17.57 16.88 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 152.6 on 3090 degrees of freedom
Multiple R-squared: 0.08439, Adjusted R-squared: 0.08409
F-statistic: 284.8 on 1 and 3090 DF, p-value: < 2.2e-16cor.test(x= covid_vote_5_no_zero$percent_trump, y= covid_vote_5_no_zero$covid_death_rate)
Pearson's product-moment correlation
data: covid_vote_5_no_zero$percent_trump and covid_vote_5_no_zero$covid_death_rate
t = 16.876, df = 3090, p-value < 2.2e-16
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.2578920 0.3224492
sample estimates:
cor
0.2905011 #Creating a new variable, whether trump or clinton was the majority vote
covid_vote_5_no_zero<- covid_vote_5_no_zero%>%
mutate(majority= case_when(percent_trump > 0.5 ~ "Trump_favor",
percent_trump < 0.5 ~ "Clinton_favor"))
head(covid_vote_5_no_zero)# A tibble: 6 × 11
county_…¹ STNAME CTYNAME covid…² perce…³ over6…⁴ Party med_i…⁵ tot_pop Rural…⁶
<chr> <chr> <chr> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
1 01001 Alaba… Autaug… 376. 0.728 0.160 Repu… 67565 55869 2
2 01003 Alaba… Baldwi… 300. 0.765 0.210 Repu… 71135 223234 3
3 01005 Alaba… Barbou… 381. 0.521 0.197 Repu… 38866 24686 6
4 01007 Alaba… Bibb C… 447. 0.764 0.167 Repu… 50907 22394 1
5 01009 Alaba… Blount… 398. 0.893 0.187 Repu… 55203 57826 1
6 01011 Alaba… Bulloc… 515. 0.242 0.169 Repu… 33124 10101 6
# … with 1 more variable: majority <chr>, and abbreviated variable names
# ¹county_fipsNEW, ²covid_death_rate, ³percent_trump, ⁴over65_pct,
# ⁵med_income_2020, ⁶Rural_urban_continuum_code_2013#creating a boxplot to compare means of two groups
ggplot(na.omit(covid_vote_5_no_zero), aes(x=majority, y=covid_death_rate))+geom_boxplot()+labs(x="Majority", y="COVID-19 Deaths per 100,000 People", title= "COVID-19 Death Rates in Clinton and Trump Majority Counties")+theme_light()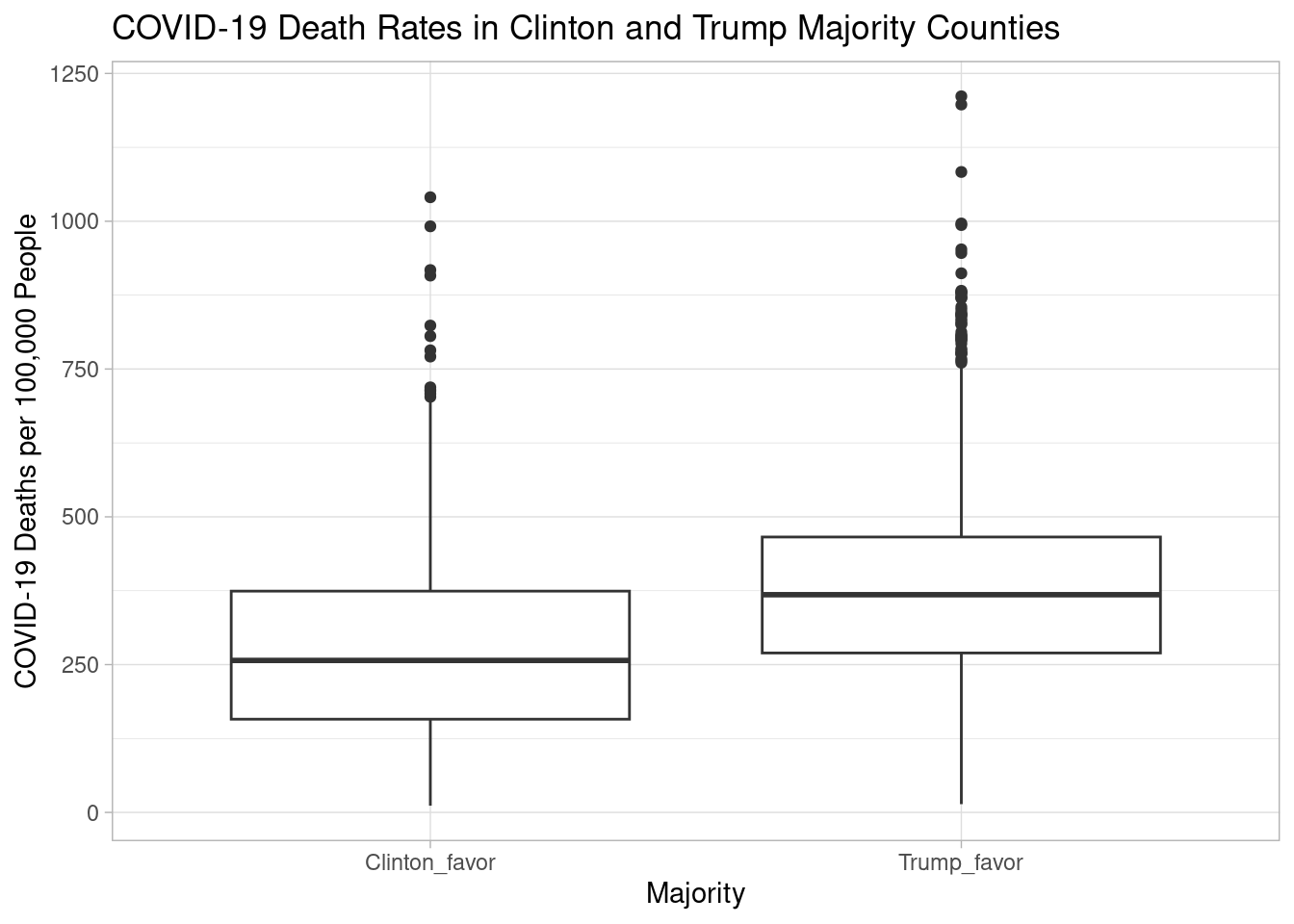
#Conducting a T test for difference of means:
t.test(covid_death_rate ~ majority, data = covid_vote_5_no_zero)
Welch Two Sample t-test
data: covid_death_rate by majority
t = -11.74, df = 858.72, p-value < 2.2e-16
alternative hypothesis: true difference in means between group Clinton_favor and group Trump_favor is not equal to 0
95 percent confidence interval:
-102.43835 -73.09319
sample estimates:
mean in group Clinton_favor mean in group Trump_favor
287.9746 375.7404 #calculating medians for both groups
#Trump favor median
filter(covid_vote_5_no_zero, majority=="Trump_favor") %>%
summarize(median(covid_death_rate, na.rm=T))# A tibble: 1 × 1
`median(covid_death_rate, na.rm = T)`
<dbl>
1 368.#Clinton favor median
filter(covid_vote_5_no_zero, majority=="Clinton_favor") %>%
summarize(median(covid_death_rate, na.rm=T))# A tibble: 1 × 1
`median(covid_death_rate, na.rm = T)`
<dbl>
1 257.The Pearson’s correlation coefficient for this simple linear model is 0.29 with an R-squared of 0.084 and a p-value below 0.01, indicating a statistically significant relationship between political partisanship and COVID-19 deaths in U.S. counties. However, because the R-squared is so low, there are likely other variables beyond political partisanship that contribute to differential COVID-19 deaths. This relationship is also statistically significant when the percent Trump variable is converted to a dummy variable (Clinton-favoring or Trump-favoring counties). A Welch Two-Sample T-test reveals a significant difference in means between Trump-favoring, and Clinton-favoring counties with a p-value below 0.01. The 95% confidence interval for this difference in death rates between Trump- and Clinton-majority counties is [-102.43835, -73.09319], with Trump-majority counties having a higher median death rate. For Clinton-majority counties, the median death rate is 257 per 100,00 people. For Trump-majority counties, the median death rate is 368 per 100,000 people.
Modeling this Relationship
Below I test a few models. The first is all variables but no transformations. The second is a log transformation of the income variable which makes sense given the possibility of diminishing returns. In other words, the decrease in COVID-19 deaths as income increases is smaller (smaller decrease) as income gets bigger (diminishing returns). The last model removes the governor party variable to see if the coefficient for percent_trump is still significant when the policy dummy variable is removed (potential multicollinearity).
#no transformations
summary(lm(covid_death_rate ~ percent_trump + over65_pct + med_income_2020 + Party+ Rural_urban_continuum_code_2013, data= covid_vote_5_no_zero))
Call:
lm(formula = covid_death_rate ~ percent_trump + over65_pct +
med_income_2020 + Party + Rural_urban_continuum_code_2013,
data = covid_vote_5_no_zero)
Residuals:
Min 1Q Median 3Q Max
-421.65 -81.69 -4.04 77.25 765.28
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 5.154e+02 2.013e+01 25.602 < 2e-16 ***
percent_trump 1.645e+02 1.751e+01 9.393 < 2e-16 ***
over65_pct -3.150e+01 5.871e+01 -0.537 0.592
med_income_2020 -4.765e-03 1.939e-04 -24.579 < 2e-16 ***
PartyRepublican 2.869e+01 5.158e+00 5.561 2.91e-08 ***
Rural_urban_continuum_code_2013 5.097e-01 1.145e+00 0.445 0.656
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 135.6 on 3085 degrees of freedom
(1 observation deleted due to missingness)
Multiple R-squared: 0.2774, Adjusted R-squared: 0.2762
F-statistic: 236.9 on 5 and 3085 DF, p-value: < 2.2e-16fit0<- lm(covid_death_rate ~ percent_trump + over65_pct + med_income_2020 + Party+ Rural_urban_continuum_code_2013, data= covid_vote_5_no_zero)
#log income
summary(lm(covid_death_rate ~ percent_trump + over65_pct + log(med_income_2020) + Party+ Rural_urban_continuum_code_2013, data= covid_vote_5_no_zero))
Call:
lm(formula = covid_death_rate ~ percent_trump + over65_pct +
log(med_income_2020) + Party + Rural_urban_continuum_code_2013,
data = covid_vote_5_no_zero)
Residuals:
Min 1Q Median 3Q Max
-438.24 -79.98 -3.49 74.02 745.66
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3663.8148 132.3950 27.673 < 2e-16 ***
percent_trump 187.1206 17.2389 10.855 < 2e-16 ***
over65_pct -10.1550 57.7624 -0.176 0.860
log(med_income_2020) -313.9957 11.7492 -26.725 < 2e-16 ***
PartyRepublican 26.2591 5.0894 5.160 2.63e-07 ***
Rural_urban_continuum_code_2013 -0.9974 1.1395 -0.875 0.381
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 133.7 on 3085 degrees of freedom
(1 observation deleted due to missingness)
Multiple R-squared: 0.2983, Adjusted R-squared: 0.2972
F-statistic: 262.3 on 5 and 3085 DF, p-value: < 2.2e-16fitlog<-lm(covid_death_rate ~ percent_trump + over65_pct + log(med_income_2020) + Party+ Rural_urban_continuum_code_2013, data= covid_vote_5_no_zero)
#log income model with policy removed (to check for possible multicollinearity with percent Trump)
summary(lm(covid_death_rate ~ percent_trump + log(med_income_2020) + over65_pct + Rural_urban_continuum_code_2013, data= covid_vote_5_no_zero))
Call:
lm(formula = covid_death_rate ~ percent_trump + log(med_income_2020) +
over65_pct + Rural_urban_continuum_code_2013, data = covid_vote_5_no_zero)
Residuals:
Min 1Q Median 3Q Max
-428.10 -83.29 -4.60 75.23 740.56
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3767.405 131.308 28.691 <2e-16 ***
percent_trump 207.212 16.795 12.338 <2e-16 ***
log(med_income_2020) -322.377 11.673 -27.617 <2e-16 ***
over65_pct -57.833 57.262 -1.010 0.313
Rural_urban_continuum_code_2013 -1.090 1.144 -0.953 0.341
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 134.2 on 3087 degrees of freedom
Multiple R-squared: 0.2924, Adjusted R-squared: 0.2915
F-statistic: 318.9 on 4 and 3087 DF, p-value: < 2.2e-16fitlog_no_policy<-lm(covid_death_rate ~ percent_trump + log(med_income_2020) + over65_pct + Rural_urban_continuum_code_2013, data= covid_vote_5_no_zero)Once the policy dummy variable is removed, the coefficients and adjusted R-squared do not change significantly so we do not need to be super concerned about the policy dummy introducing multicollinearity.
Diagnostic Plots:
#fit0
par(mfrow= c(2,3)); plot(fit0, which=1:6)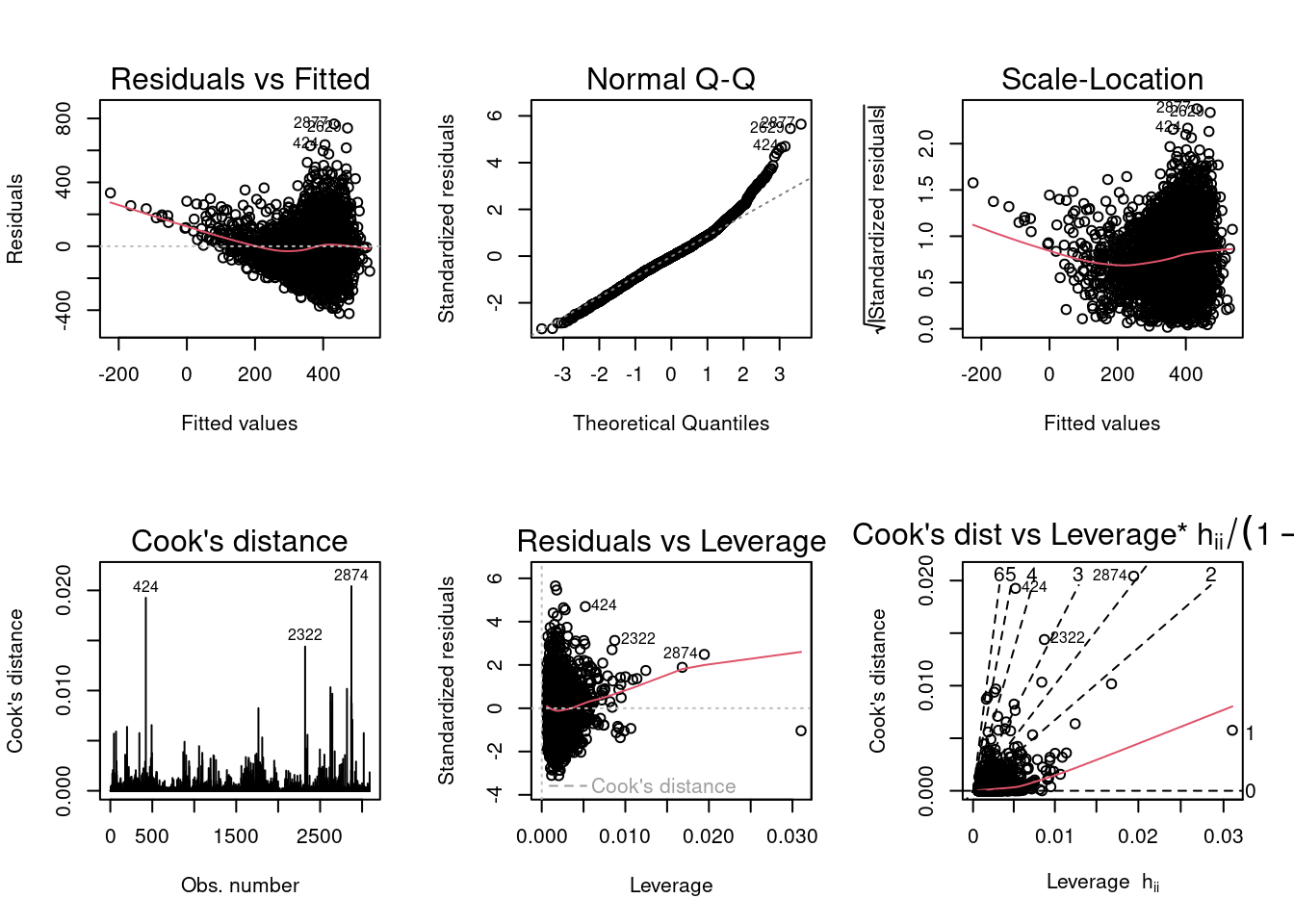
#fitlog
par(mfrow= c(2,3)); plot(fitlog, which=1:6)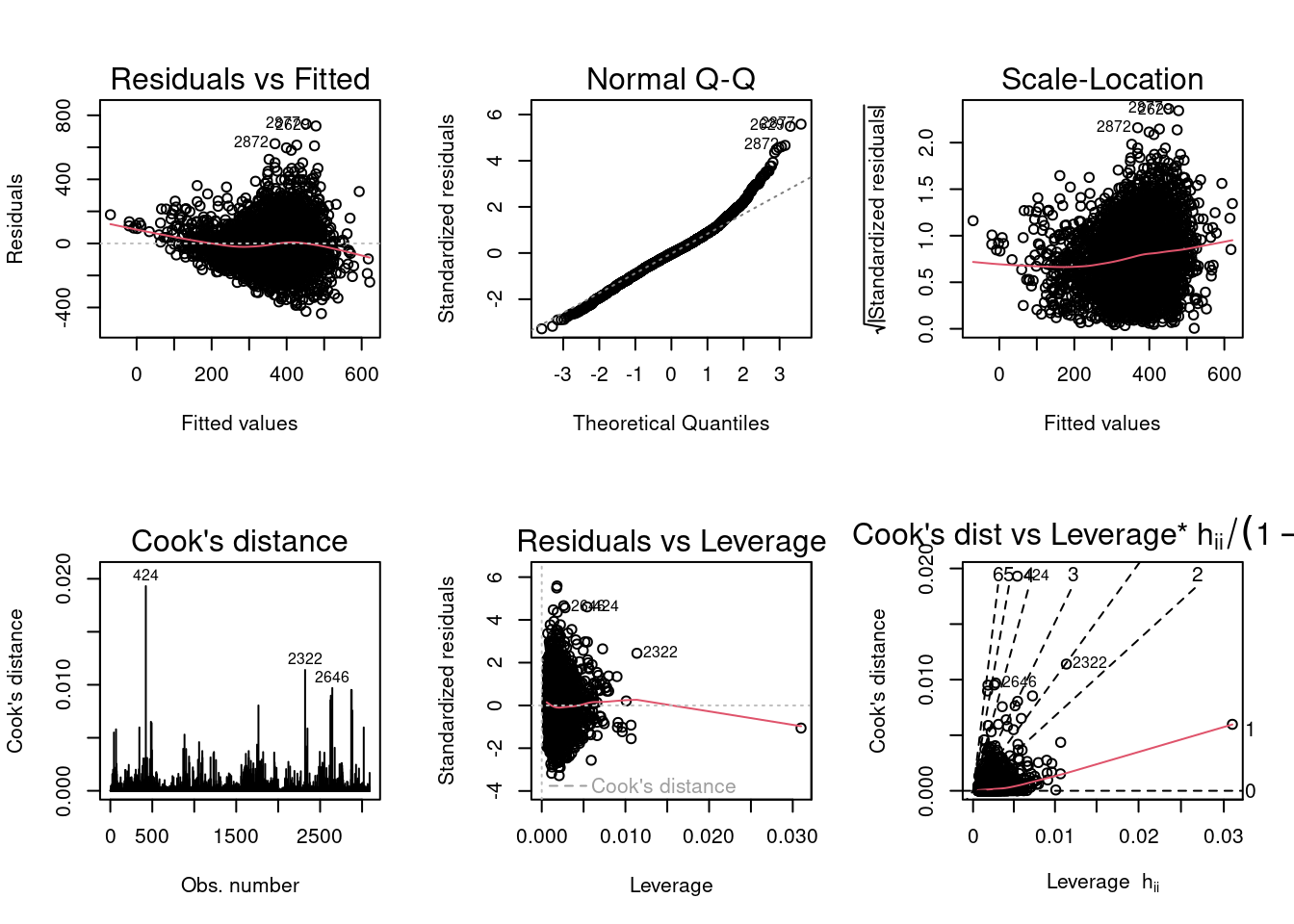
According to the diagnostic plots for fitlog (the second set of plots above where where covid_death_rate is log transformed,) the residuals seem to have a slight trend (higher fitted values have larger residuals) but no funnel shape so constant variance is not violated. Again, for the Q-Q, plot, higher theoretical quantiles have larger standardized residuals (there are high residuals at higher values), however the trend is not overly concerning. The scale location graph does not look too bad, the line is approximately horizontal. For the Cook’s distance plot, there may be a couple outliers affecting the model according to 4/n which is approximately 0.0013, however there a no points with a Cook’s distance larger than 1. Residuals versus leverage and Cook’s distance versus leverage plots look okay, there does not appear to be any observations with both high leverage and Cooks distance. The plots for the log transformed income variable look better than the plots without the transformation.
#Below the models are displayed in regression table for ease of comparison.
stargazer(fit0, fitlog, fitlog_no_policy, type= "text", style="apsr", covariate.labels=c("Percent Trump", "Percent Over 65", "Median HH Income", "log(Median HH Income)", "Policy- Republican", "Population Density Code", "Percent Vaccinated"), dep.var.labels= "COVID-19 Death Rate")
-----------------------------------------------------------------------------------------------------
COVID-19 Death Rate
(1) (2) (3)
-----------------------------------------------------------------------------------------------------
Percent Trump 164.471*** 187.121*** 207.212***
(17.511) (17.239) (16.795)
Percent Over 65 -31.503 -10.155 -57.833
(58.709) (57.762) (57.262)
Median HH Income -0.005***
(0.0002)
log(Median HH Income) -313.996*** -322.377***
(11.749) (11.673)
Policy- Republican 28.685*** 26.259***
(5.158) (5.089)
Population Density Code 0.510 -0.997 -1.090
(1.145) (1.139) (1.144)
Percent Vaccinated 515.423*** 3,663.815*** 3,767.405***
(20.132) (132.395) (131.308)
N 3,091 3,091 3,092
R2 0.277 0.298 0.292
Adjusted R2 0.276 0.297 0.291
Residual Std. Error 135.634 (df = 3085) 133.655 (df = 3085) 134.200 (df = 3087)
F Statistic 236.867*** (df = 5; 3085) 262.342*** (df = 5; 3085) 318.928*** (df = 4; 3087)
-----------------------------------------------------------------------------------------------------
*p < .1; **p < .05; ***p < .01 PRESS(fit0)Error in PRESS(fit0): could not find function "PRESS"PRESS(fitlog)Error in PRESS(fitlog): could not find function "PRESS"PRESS(fitlog_no_policy)Error in PRESS(fitlog_no_policy): could not find function "PRESS"In both models, the coefficient of interest percent_trump, is positive and significant at the 0.001 significance level. These models suggests that there is evidence that counties that vote in greater numbers for Trump experience higher COVID-19 death rates. The PRESS statistic for the second model (where income is logged) is the lowest, indicating this may be a better model than without the transformation.
Adding a Control for Vaccines
Data from the CDC (2022) on vaccination rates was added to the model. Vaccination rates as of March 19, 2022 is taken (same as the cut-off date for the COVID-19 deaths outcome variable) and is expressed as a proportion of the population fully-vaccinated (1 dose of Janssen or 2 doses of the Pfizer or Moderna vaccine) (CDC 2022).
#this is the code I used to reduce the file size of the vaccine data to make it upload-able to Google Sheets. I used the proportion of the population in the county with a complete primary series of vaccine(s) as of 3/19/2022 (same as cut off date for COVID-19 death data)
#vaccines<-read_csv("COVID-19_Vaccinations_in_the_United_States_County (1).csv")
#vaccines<-filter(vaccines, Date =="03/19/2022")
#vaccines<-rename(vaccines, "county_fipsNEW"=FIPS)
#vaccines<-select(vaccines, "county_fipsNEW", Series_Complete_Pop_Pct)
vaccines<-read_sheet("https://docs.google.com/spreadsheets/d/18X8jPAatcAxE1-kuadH4FB44Sp2Zpg6Q-kw8efqYY-g/edit?usp=sharing")
head(vaccines)# A tibble: 6 × 2
county_fipsNEW Series_Complete_Pop_Pct
<chr> <chr>
1 38053 21.9
2 48181 44.7
3 06081 83.3
4 31079 48.7
5 24023 49.3
6 48057 57.1 #joining to the df
covid_vote_6<- covid_vote_5_no_zero %>%
left_join(vaccines, by= "county_fipsNEW")
#converting Series_Complete_Pop_Pct to numeric
covid_vote_6$Series_Complete_Pop_Pct<-as.numeric(covid_vote_6$Series_Complete_Pop_Pct)
#converting percent vaccinated to proportion
covid_vote_6$fullvax_prop<-covid_vote_6$Series_Complete_Pop_Pct/100
head(covid_vote_6)# A tibble: 6 × 13
county_…¹ STNAME CTYNAME covid…² perce…³ over6…⁴ Party med_i…⁵ tot_pop Rural…⁶
<chr> <chr> <chr> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
1 01001 Alaba… Autaug… 376. 0.728 0.160 Repu… 67565 55869 2
2 01003 Alaba… Baldwi… 300. 0.765 0.210 Repu… 71135 223234 3
3 01005 Alaba… Barbou… 381. 0.521 0.197 Repu… 38866 24686 6
4 01007 Alaba… Bibb C… 447. 0.764 0.167 Repu… 50907 22394 1
5 01009 Alaba… Blount… 398. 0.893 0.187 Repu… 55203 57826 1
6 01011 Alaba… Bulloc… 515. 0.242 0.169 Repu… 33124 10101 6
# … with 3 more variables: majority <chr>, Series_Complete_Pop_Pct <dbl>,
# fullvax_prop <dbl>, and abbreviated variable names ¹county_fipsNEW,
# ²covid_death_rate, ³percent_trump, ⁴over65_pct, ⁵med_income_2020,
# ⁶Rural_urban_continuum_code_2013#adding vaccines to the model
summary(lm(covid_death_rate ~ percent_trump + over65_pct + log(med_income_2020) + Party+ Rural_urban_continuum_code_2013 + fullvax_prop, data= covid_vote_6))
Call:
lm(formula = covid_death_rate ~ percent_trump + over65_pct +
log(med_income_2020) + Party + Rural_urban_continuum_code_2013 +
fullvax_prop, data = covid_vote_6)
Residuals:
Min 1Q Median 3Q Max
-439.05 -80.24 -2.91 74.22 733.43
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3597.208 133.535 26.938 < 2e-16 ***
percent_trump 182.939 21.952 8.334 < 2e-16 ***
over65_pct 23.667 58.391 0.405 0.685
log(med_income_2020) -307.942 12.374 -24.886 < 2e-16 ***
PartyRepublican 27.043 5.071 5.333 1.04e-07 ***
Rural_urban_continuum_code_2013 -1.042 1.129 -0.923 0.356
fullvax_prop -8.208 29.505 -0.278 0.781
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 131.8 on 3062 degrees of freedom
(23 observations deleted due to missingness)
Multiple R-squared: 0.3003, Adjusted R-squared: 0.299
F-statistic: 219.1 on 6 and 3062 DF, p-value: < 2.2e-16fitvaccines<-(lm(covid_death_rate ~ percent_trump + over65_pct + log(med_income_2020) + Party+ Rural_urban_continuum_code_2013 + fullvax_prop, data= covid_vote_6))
stargazer(fit0, fitlog, fitvaccines, type= "text", style="apsr", covariate.labels=c("Percent Trump", "Percent Over 65", "Median HH Income", "log(Median HH Income)", "Policy- Republican", "Population Density Code", "Percent Vaccinated"), dep.var.labels= "COVID-19 Death Rate")
-----------------------------------------------------------------------------------------------------
COVID-19 Death Rate
(1) (2) (3)
-----------------------------------------------------------------------------------------------------
Percent Trump 164.471*** 187.121*** 182.939***
(17.511) (17.239) (21.952)
Percent Over 65 -31.503 -10.155 23.667
(58.709) (57.762) (58.391)
Median HH Income -0.005***
(0.0002)
log(Median HH Income) -313.996*** -307.942***
(11.749) (12.374)
Policy- Republican 28.685*** 26.259*** 27.043***
(5.158) (5.089) (5.071)
Population Density Code 0.510 -0.997 -1.042
(1.145) (1.139) (1.129)
Percent Vaccinated -8.208
(29.505)
Constant 515.423*** 3,663.815*** 3,597.208***
(20.132) (132.395) (133.535)
N 3,091 3,091 3,069
R2 0.277 0.298 0.300
Adjusted R2 0.276 0.297 0.299
Residual Std. Error 135.634 (df = 3085) 133.655 (df = 3085) 131.815 (df = 3062)
F Statistic 236.867*** (df = 5; 3085) 262.342*** (df = 5; 3085) 219.078*** (df = 6; 3062)
-----------------------------------------------------------------------------------------------------
*p < .1; **p < .05; ***p < .01 Even once we control for vaccination rates, the percent Trump coefficient is still positive and significant (though to a slightly lesser magnitude). This suggests that vaccine uptake is responsible for a part of this difference however, other health behaviors may contribute to higher COVID-19 death rates in pro-Trump counties. Previous research has attributed this difference mainly to vaccine uptake (Wood and Brumfiel 2021), however, this model shows there are more variables at play, and that differential death rates in pro-Trump counties are still significant when we control for vaccine uptake (though to a lesser magnitude). One limitation of this variable is that a majority of deaths likely occurred prior to when the vaccine was available- which may explain why while the coefficient is negative, it is not statistically significant in our model.
References
Ballotpedia. (2022). Partisan composition of governors. Accessed [November 10, 2022]. https://ballotpedia.org/Partisan_composition_of_governors
Centers for Disease Control. (2022). COVID-19 Vaccinations in the United States,County. Accessed [December 6, 2022]. https://data.cdc.gov/Vaccinations/COVID-19-Vaccinations-in-the-United-States-County/8xkx-amqh
Economic Research Service. (2022). Unemployment and median household income for the U.S., States, and counties, 2000-2021. Accessed from the United States Department of Agriculture [November 10, 2022]. https://www.ers.usda.gov/data-products/county-level-data-sets/county-level-data-sets-download-data/
Jones, B. (2022). The Changing Political Geography of COVID-19 Over the Last Two Years. Pew Research Center. March 3, 2022. https://www.pewresearch.org/politics/2022/03/03/the-changing-political-geography-of-covid-19-over-the-last-two-years/
Mayo Clinic Staff. (2022). COVID-19: Who’s at Higher Risk of Serious Symptoms? Accessed from Mayo Clinic, [November 11, 2022]. https://www.mayoclinic.org/diseases-conditions/coronavirus/in-depth/coronavirus-who-is-at-risk/art-20483301
MIT Election Data and Science Lab. (2021) County Presidential Election Returns 2000-2020. Accessed from the Harvard Dataverse [October 11, 2022]. https://doi.org/10.7910/DVN/VOQCHQ
National Center for Health Statistics. (2022). Provisional COVID-19 Deaths by County, and Race and Hispanic Origin. Accessed from the Centers for Disease Control [October 11, 2022]. https://data.cdc.gov/d/k8wy-p9cg
USA Facts. (2022). US COVID-19 cases and deaths by state. Accessed [November 10, 2022]. https://usafacts.org/visualizations/coronavirus-covid-19-spread-map/?utm_source=usnews&utm_medium=partnership&utm_campaign=2020&utm_content=healthiestcommunitiescovid
United States Census Bureau. (2019). Annual County Resident Population Estimates by Age, Sex, Race, and Hispanic Origin: April 1, 2010 to July 1, 2019 (CC-EST2019-ALLDATA). Accessed from Census.gov [November 11, 2022].https://www.census.gov/data/tables/time-series/demo/popest/2010s-counties-detail.html
Wood, D. and Brumfiel, G. (2021). Pro-Trump counties now have far higher COVID death rates. Misinformation is to blame. NPR. December 5, 2021. https://www.npr.org/sections/health-shots/2021/12/05/1059828993/data-vaccine-misinformation-trump-counties-covid-death-rate